
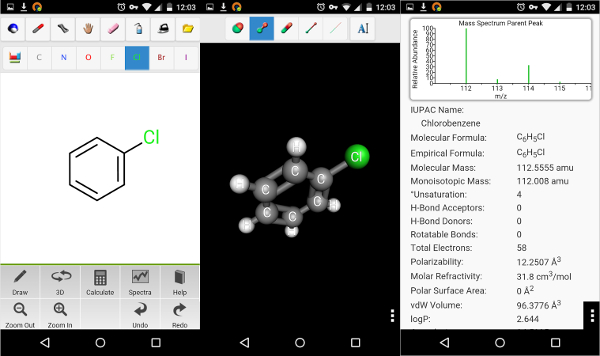
Stoichiometric and additive naming, UTF8 text format output, a lot of new trivial name inclusions and PIN (Preferred IUPAC Name) in addition to Traditional IUPAC naming, more nomenclature systems including thorough Improperly drawn rectangular geometry warnings are no longer flagged in rings of size 3 or 4.ĬhemDoodle 2D v11.12.0 is a feature update with a focus on IUPAC naming.A rounding error occured when rendering 4 membered rings with double bonds in certain style sheets, leading to only one side being fit correctly, this has been resolved.Lewis dot structures were incorrectly output with 60 degree angles for certain atom centers with 4 substituents, this has been rectified.Fixed issue where charges were no longer rendered after using the graph reduction functions.Corrected issue where in certain cases involving double bonds in multiple equivalent rings, the second bond line would be locked in place and could not be flipped to the other side.Minor improvements to JDX spectra input and much faster output to JDX format for simulated NMR spectra.Improvements to congested spiro structure layout.Significant speed improvements for generating isotopic distributions.This option is found in the Preferences window, under the Functions tab in the Stereochemistry section. We now include a second type of IUPAC name output, an Attempted Preferred IUPAC Name (PIN) inĪddition to the Traditional IUPAC name.IUPAC naming has been upgraded, please see the updated chapter 15 of the ChemDoodle User Guide for detailed instructions.Additionally, several issues have been corrected, described below. Traditional IUPAC names will be modified by the options inĬhemDoodle, while the PIN will not be. ChemDoodle will try its best to generate the PIN, but is not Guaranteed to, which is why it is called an Attempted PIN. Name menu item will now display both name types. Auto-updating labels will display theĪttempted PIN, but can be changed to show the Traditional IUPAC name instead in theįunctions panel of the Preferences window. ChemDoodle will attempt to intelligently name your More nomenclature systems, including stoichometric and additive naming for polyatomic or.Of the ChemDoodle User Guide for further explanation. sy2), Tripos Sybyl Line Notation (.sln), Beilstein ROSDAL (.ros), XYZ Files (.Structures as clearly as possible using the best nomenclature system, and will respect zero-orderīonds when separating structures. mmod), Schrödinger Maestro (.mae), Standard Molecular Data (.smd), Tripos Mol2 (.mol2. ent), RCSB Protein Data Bank Markup Language (.xml. mmcif), RCSB MacroMolecular Transmission Format (.mmtf), RCSB Protein Data Bank Files (.pdb. rd), MDL RXNFiles, both V2000 and V3000 connection tables (.rxn), MMI SketchEl Molecule (.el), Molinspiration JME String (.jme), RCSB Binar圜IF (.bcif), RCSB Macromolecular Crystallographic Information File (.cif. dx), ISIS Sketch File (.skc), ISIS Sketch Transportable Graphics File (.tgf), MDL MOLFiles, both V2000 and V3000 connection tables (.mol. com), IUPAC InChI (.inchi), IUPAC JCAMP-DX (.jdx. Read and write many popular chemical file types for working with the applications you use:ĪCD/ChemSketch Documents (.sk2), ChemDoodle Documents (.icl), ChemDoodle 3D Scenes (.ic3), ChemDoodle Javascript Data (.cwc.js), CambridgeSoft ChemDraw Exchange (.cdx), CambridgeSoft ChemDraw XML (.cdxml), Crystallographic Information Format (.cif), CHARMM CARD File (.crd), ChemAxon Marvin Document (.mrv), Chemical Markup Language (.cml), Daylight SMILES (.smi. Algorithmic Analysis of Cahn−Ingold−Prelog Rules of Stereochemistry: Proposals for Revised Rules and a Guide for Machine Implementation. and is 100% accurate in all 300 test cases provided. The CIP algorithm in ChemDoodle is validated against the test suite provided by Hanson et. Stereochemical features in your structures will be assigned "R", "S", "E", "Z", "M" and "P" descriptors.

to remove any ambiguities and describe a completely consistent system for CIP assignments.ĬhemDoodle implements all 6 current CIP rules as well as auxilliary desciptors and mancude ring support. The most recent CIP rules from IUPAC were then algorithmically analyzed and standarized by Hanson et al.
These rules were adopted by IUPAC for naming standards and fully described in the Blue books.

While flawed, they have seen many revisions over the decades and were clarified by the work of Paulina Mata. The CIP rules have long been the standard for describing configurations of stereochemical features in a molecule.


 0 kommentar(er)
0 kommentar(er)
